1 - Cocoa Research Unit, University of the West Indies, St. Augustine, Trinidad and Tobago
Genetic diversity is Currently assessed In the ICG,T using Isozyme Electrophoresis and RAPD. The information obtained from this assessment is used for establishing a “working collection” within ICG,T Containing a reduced number of clones with large genetic diversity and interesting agronomiC traits. Proposals are made to integrate the results of this assessment in to the Cocoa breeding strategy.
Introduction
The Cocoa Research Unit (CRU) is in charge of the maintenance, description and evaluation of the 2,300 cocoa clones present in the International Cocoa Genebank (ICG,T) in Trinidad. Besides the morphological description of the clones, biochemical (Isozyme Electrophoresis) and molecular (RAPD) markers are currently used to characterise these clones and to assess the level of genetic diversity present in the
ICG,T.
Material and methods
Plant material
Methods
RAPD. DNA is extracted according to the method proposed by Edwards et at (1991), as modified by Johnson et a!. (1992). Amplifications are carried out following the method described by Christopher and Sounigo (1996). Thirteen operon primers are used to generate 30 polymorphic reproducible and scorable markers for these analyses. The amplification products are run on a 1.6% agarose gel and visualised under ultra violet light after staining with ethidium bromide.
Statistical analyses: The genetic diversity is measured using multivariate analyses. The level of genetic diversity within populations is measured by the Shannon index (Magurran 1988) when RAPO data are analysed. For isozyme data, the mean number of polymorphic markers, the mean number of alleles per locus and levels of observed and expected heterozygosity are calculated. The genetic relationships between populations are visualised using cluster analyses performed on Rogers-Wright distances (Wright 1978) in the case of RAPD data, and on Nei distances (Nei 1972) in the case of isozymes, calculated on pairs of populations.
The results and their use
Characterization using isozymes
The isozyme characterisation data for the clones are included in the International Cocoa Germplasm Database (ICGD) (Wadsworth et at 1997). These can be used by all cocoa breeders for:
Level of genetic diversity within populations
The levels of diversity observed in some of the populations using the RAPD technique are indicated in Table 1. The information about the genetic origin of the clones (number of mother trees) was obtained from the ICGD (Wadsworth et at1997).The levels of genetic diversity observed are not significantly correlated to the sample sizes of these populations indicating that the differences are not simply caused by bias arising from differences in sampling. The results show that the highest levels of diversity where found in populations that supposedly originate from the lowest numbers of mother trees (SCA, MC and MO), which is rather surprising. This could be explained in the case of MO by the hypothesis made by Bartley (1993) that genotypes from different geographic origins could have been named MO by mistake. Even the accessions from the SPA population, supposedly originating from a single pod, present a level of genetic diversity almost equal to the one shown by the NA population, which is expected to originate from 14 mother trees. The populations from French Guiana present a low level of genetic diversity, except ELP, which presents a fair level of diversity, equal to the one presented by the NA population.
Relationship between populations
Conclusion
Isozyme and RAPD marker techniques currently provide us with useful information on the genetic diversity existing in the ICG,T. However, these two types of markers suffer from some drawbacks:
The introduction of another technique, especially one using Co-dominant markers, would therefore be desirable. PCR-based SSR markers, developed for cocoa by Lanaud eta!. (1999) could be a technique of choice, once the necessary equipment for sequencing gels is available at CRU.
References
Bartley B.G.D. 1993. Notes on the meaning and origins of clone names. Personal communication to Michelle End, The University of Reading, UK

Biotech Glossary |
Bioinformatics |
Lab Protocol |
Notes |
Malaysia University |
Evaluation and Use of the Genetic Diversity Present in the International Cocoa Genebank (ICG,T), in Trinidad
Olivier Sounigo2, Yvette Christopher’, Stacy Ramdahin’, RoMina Unaaharan’ and Antoinette Sankar
2 - Cirad-Cp, TA 80/02, Avenue Agropolis, 34098 Montpellier Cedex 5, France, Attached to the Cocoa Research Unit
The ICG,T is composed of:
Isozyme E/ectrophoresis. Five systems are currently used, following the methodology of Lanaud (1986): ACP, ADH, IDH, MDH and GPI.
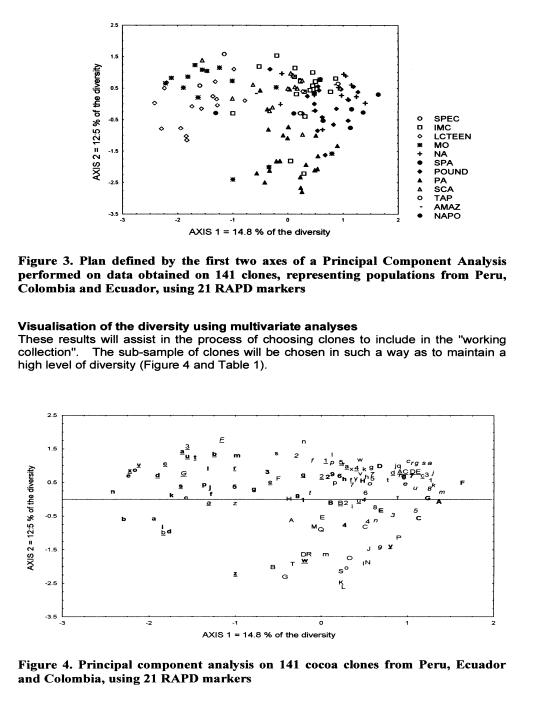
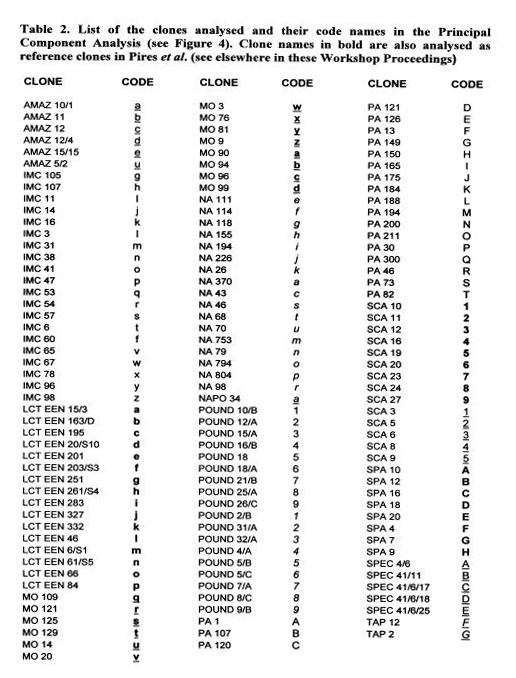
These data are currently used for the establishment of a ‘working collection for the CFC/ICCO/IPGRI project on Cocoa Germplasm Utilization and Conservation’ (Sounigo et at 2000). A sub-sample of the most useful clones in the ICG,T is selected for inclusion in this ‘working collection”. The “usefulness” is defined in terms of resistance to diseases and favourable pod and bean characteristics. In order to maintain a high level of diversity in the working collection, an attempt was made to represent a large number of populations, the level of representation of each population depending on the level of genetic diversity it contains.
.jpg)
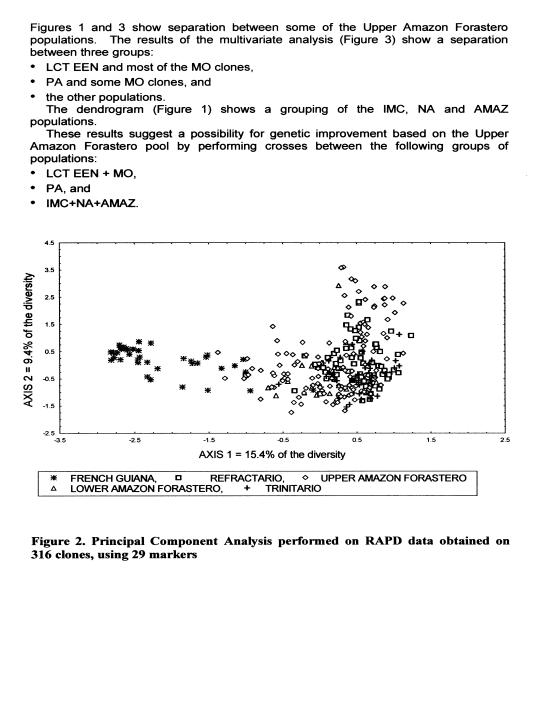
The dendrogram (Figure 1) and the plan defined by the first two axes of a multivariate analysis (Figure 2) clearly show a separation between the populations from French Guiana and the other populations. This result shows the usefulness of cocoa types from this region for the enrichment of the cocoa germplasm collections. The unique nature of this material makes it imperative to evaluate its combining ability with parents representing other populations. This type of study has already started at Cirad French Guiana, but as yet only with clones collected on the banks of the Camopi river. This study should be completed with the evaluation of clones collected on the banks of the other rivers, especially along the Eulepousing river since this population shows the highest level of diversity.
.jpg)
Christopher V., 0. Sounigo. 1996. The use of RAPD for characterization and genetic diversity assessment of cacao. Pages 38-51 in Annual report of the Cocoa Research Unit for 1995. The Cocoa Research Unit, St. Augustine, Trinidad,
Edwards K., C. Johnstone and C, Thompson. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nod. Acid. Res., 19: 1349.
Johnson E., JR. Russell, F. Hosein, W, Powell and P. Waugh. 1992. A laboratory manual for RAPD analyses in cocoa. In internal report of the Cocoa Research Unit (unpublished).
Lanaud C. 1986. Utilisation des marqueurs enzymatiques pour ‘etude genetique do cacaoyer:
Theobroma cacao L. I. ContrOle genetique et linkage’ de neuf marqueurs enzymatiques. Cafe Cacao The 30: 259-270.
Lanaud C., AM. Risterucci, I. Pieretti, M. Faique. A. Bouet and P.J.L. Lagoda. 1999. Isolation and characterization of microsatellites in Theobroma cacao L. Mol. Ecol. 12(8): 2141.
Magurran A.E. 1988. Ecological diversity and its measurement. Croom Helm. London. 179 pp.
Nei M. 1978. Genetic distance between populations. Amer. Naturalist. 106: 283-292.
Sounigo 0., V. Mooleedhar, D. Iwaro, F. Bekele, TN. Sreenivasan, J.M. Thevenin, N. Khan and
D. Butler. 2000. Strategy to establish a CFC Project Collection. Pages 29-37 in
Proceedings of the CFC/ICCO/IPGRI Project Workshop. 1-6 February 1998. Montpellier.
France. IPGRI.Rome, Italy.
Wadsworth R.M., CS. Ford, M.J. End and P. Hadley (eds). 1997. International cocoa germplasm database. The London International Financial Futures and Options Exchange (LIFFE), London, UK.
Wright S. 1978. Evolution and the genetics of populations. vol 4. Variability within and among natural populations. University of Chicago Press,Chicago, U.S.A.