Cocoa swollen shoot virus disease (CSSVD) is one of the most enduring features of cocoa production in Ghana in particu’ar, and West Africa in general. First recorded in 1938 in the Eastern region of the then Gold Coast, CSSVD is now present in virtually all cocoa growing areas of Ghana. as well as in neighbouring Togo, Nigeria, Cameroon and, possibly, in Cote divoire. The virus is indigenous to West Africa, present in forest trees and appears to have transferred to cocoa following the introduction of the crop. Crop losses due to the disease have affected the economies of countries like Ghana for which cocoa is an import export commodity. It has caused changes to the way of life of whole communities as they are forced to migrate, and the environment as forests are cleared for new farms. The prime control practice has been based on a “zero tolerance” philosophy which required that all infected trees and those in contact with them be removed. In Ghana, this has resulted in the destruction of millions of trees during the last 50 years, and many millions more are awaiting removal. With new farms getting rapiWy infected, breeding for disease resistance has become imperative. Developments in biotechnology research have provided new tools to aid cacao breeders. These include the use of micropropagation techniques, particularly somatic embryogenesis, to produce large numbers of plants with identical genetic background, coupled with transformation protocols for cocoa transformation. Another is the development of new methods of virus inoculation and diagnosis, prerequisites for resistance screening. Molecular cloning methods have enabled the isolation of full-length infectious cbnes of severe isolates from Togo and Ghana. Mild isolates of the virus with potential uses in cross-protection have been isolated by the same method which enables researchers to overcome the low virus concentrations and/or low infectivity associated with them. These have been used to infect cocoa beans and young seedlings by particle bombardment and/or Agrobacterium~mediated micro-injection. With these tools, it is now possib!e to quanti& virus inoculum used in challenging new cultivars in virus disease resistance breeding programmes. New CSSV-specific primers have been designed for disease indexing by polymerase chain reaction (PCR). Cloned virus DNA or cloned virus-coded PCR DNA products have also been used in virus disease indexing as labelled probes for DNA detection by Southern (1975) or dot blot hybridisation analysis. Ultimately, it is anticipated that the combination of new methods for generating large numbers of plants for noculum delivery and virus detection wirl facilitate the deveJopment of new pest and thsease resistant varieties for sustainable cocoa production.
Importance and spread of the disease
Cocoa swollen shoot virus disease (CSSVD) was first reported in the 1930s in the eastern region of the country known then as Gold Coast, now as Ghana (Anonymous 1936a and b). Since then, it has grown in importance, constituting perhaps the single most enduring feature of cocoa production, especially in the Eastern region, previously the leading producing area in Ghana (Thresh 1991), The disease also occurs with varying degrees of severity in all cocoa producing areas of Ghana. Along the West African region, CSSVD occurs with varying degrees of importance in the Republic of Togo, Nigeria, Cameroon and Cote d’lvoire.
In West Africa, swollen shot disease is characterised by an array of host responses including transient red vein banding in very young leaves, systemic yellow mosaic,
round pods (with few deformed beans), and the stem swelling from which the disease derives its name. Severe isolates often cause death of infected trees.
Virus diseases of cocoa have also been reported from other parts of the world, including Tanzania (Zanzibar) in East Africa; Sabah Province of Malaysia, Sri Lanka, Java and Sumatra in Asia; Costa Rica, the Dominican Republic and Trinidad and Tobago in the Americas.
This paper however, is about the disease caused by a bullet shaped virus isolated from infected tissue in Ghana and the Republic of Togo. Cocoa swollen shoot virus (CSSV) was proposed as a member of the Badnavirus group (Lockhart 1990) because of its non-enveloped bacilliform particles (Brunt eta!., 1964), and its double stranded DNA genome (Lot et at 1990). Infectious clones of Togolese and Ghanaian isolates (Hagen et a!. 1993; Sackey et a!. 1995) have demonstrated that the red vein banding, systemic mosaic and stem swelling symptoms of CSSVD are due to this Badnavirus. It is reasonable to assume that the root swelling, the round pods and eventual death associated with severe CSSVD are also due to this virus, but this remains to be demonstrated.
Swollen shoot disease is a classic case of the consequences of introducing a non-native crop species, which entails risk of exposing these species to unknown local pests and diseases that were previously of no importance. Cocoa was introduced from South America during the middle to late 19th century into the forested areas of West Africa. Cocoa cultivation rapidly expanded resulting in vast, almost continuous stands beneath the remaining trees of selectively thinned forests. Early in the 20” century, vulnerability to swollen shoot virus, which the crop had never encountered before, became apparent. Infection, together with the mealybug vector came from naturally infected, widespread woody forest trees belonging to the families Bombaceae, Steroulaceae and Tiliaceae, such as baobab (Adansonia digitata), silk cotton tree (Ceiba petandra) and especially, Cola chlamydanta. In these wild indigenous hosts, there were no symptoms/disease probably because of a long association with the virus/viruses (Thresh 1980; Thresh and Owusu 1986; Thresh et al. 1988). Once the virus had established itself and becomes prevalent in cocoa, the wild host became of limited importance in further epidemic development.
In West Africa, the worst affected area has been Ghana. Following a “zero tolerance” policy, the British colonial government launched an eradication programme and by 1965 had destroyed over 100 million trees. The programme was re-designated ‘cutting out” as a control practice, perhaps in recognition of the futility of eradication”, and by 1982, 185.5 million trees had been removed, all in the Eastern region. It was estimated at that time that there were a further 31 million trees waiting to be removed (Ollennu et at 1989). A large area of the region was designated an “area of mass infection” (AMI). It was also estimated that only 23% of all infected trees in a new outbreak were identified because many of the infected trees are not noticed or are in a latent stage (Ollennu et at 1989).
The disease and its eradication programme have been costly, both in monetary terms, and through the impact on the environment and the farming communities. Economic losses were incurred by the country in terms of foreign exchange revenue, as well as the cost of implementation (labour, compensation to farmers and loss of income). The destruction of the equivalent of some 200,000 hectares of cocoa farms left many farmers destitute. In response, more resilient farmers simply moved on into new forests, leading to loss of forest cover. Abandoned farms were often victims of climatic degradation.
Disease control measures
Attempts were made to control the mealy bug vectors using contact and systemic insecticides, as well as biological control agents. So far, these efforts have yielded few
returns perhaps because of the paucity of basic information on these insects. It is known that as many as 16 species of mealybugs are involved in the transmission of CSSV. and thus biological control, for example, would require detailed studies on each. These studies are ongoing.
The World Bank in conjunction with the Government of Ghana launched two Cocoa Rehabilitation Projects in which cutting out was combined with block plantings using new cocoa introductions from the CRIG breeding programmes. The results of these initiatives have been presented elsewhere, but generally are dependent on cooperation of farmers, availability of improved (disease resistant/tolerant) planting material, and financial resources to implement the programmes.
The most persistent effort has, however, been the resistance breeding programmes in different producing countries, particularly, Ghana and logo. Since CSSV is not present in the centre of origin of the species, there is unlikely to be any specific CSSV resistance to be exploited in the wild germplasm. This means that breeding for resistance, at best, is an attempt to bring together the properties inherent in different cocoa genotypes that might create a barrier between the vector and host, or inhibit the infection process of the virus. These are often physiological characteristics of the clone, including such properties as plant tissue or sap palatability to the vector, and presence of components associated with host response (phytoalexins) to invasion by foreign substances. The main elements of the breeding programmes are therefore:
In practice. breeding requires an efficient method of inoculum delivery and methods of disease indexing. Its success is dependent on access to a wide variety of clones for screening. There are questions for which breeders need answers. For example. how do you know that if new promising genotypes are being introduced, that they are really different and that a significant reduction in virus impact can be obtained at farmers level? Could newly introduced accessions harbour some unknown pathogen that would be released into your environment? How do you challenge the new clones with the pathogen/pest of interest, and evaluate their impact in a systemic manner?
Fortunately, there are many new technologies, which can be applied to assist breeders to answer these and many other questions. These include technologies for disease indexing, and virus inoculum delivery in resistance screening for breeders.
Disease indexing
The role of the intermediary quarantine facilities is to reduce the risk of transferring pests and diseases as germplasm is exchanged between breeding programmes. For CSSV, there have been several methods of disease indexing which still have relevance. They include:
Increasingly, time is of essence and it is important to have definitive evidence of the disease status of plant tissue. Enzyme linked immunosorbent assay and related serological techniques have been used, with monoclonal and polyclonal antisera developed in collaborative research between the Cocoa Research Institute of Ghana
and the Institute for Biochemistry and Plant Virology in Brauschweig, Germany Though serological methods proved useful for disease diagnosis, the antisera available were based on the severe CSSV IA from the Eastern region of Ghana as immunogen, thus limiting them to detection of related isolates.
Over the last few years, polymerase chain reaction (PCR) has been developed for the detection of CSSV. The main thrust of this development is the detection of sub-picogram quantities of all bacilliform DNA viruses in tissues associated with infections, including latent infections. Initially, primers were designed based on nucleotide sequences derived from the severe IA isolate, and these have been used to detect several isolates of the virus in leaf tissue (Hoffmann et a!. 1997; Sackey et a!. I 990 and 1998). The current approach is to develop primers that detect a wide range of Badnaviruses, including those from bananas, plantains (BSV), sugarcanes (ScBV) and commelina (C0YMV), all of which are plant species often found in association with cocoa.
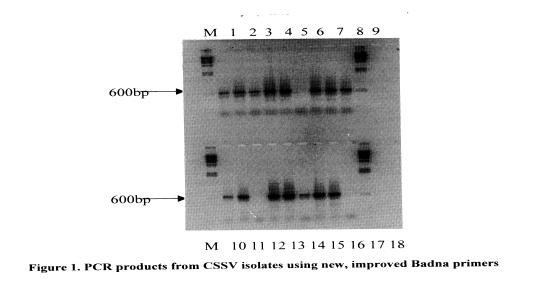
Based on this philosophy, a project was designed in which a database of nucleotide sequences from a conserved region, spanning nucleotides 5300 to 7000, of the virus genome was to be used for the design of new primers. Thirty-six isolates of CSSV from the CRIG virus museum were used in the study in which universal Badna primers designed by Lockhart and Olszewski (1993) were used to amplify virus DNA. Cloned PCR DNA amplification products were then sequenced and the data used to generate new primers. The first of the two primers incorporated sequences from severe CSSV isolates (1A, DaBV, CoYMV, BSV, KTSV) and the second was based on sequences from other severe isolates (CSSV IA and Nsaba from the central region of Ghana, CoYMV, BSV, ScBV, KTSV and DaBV).
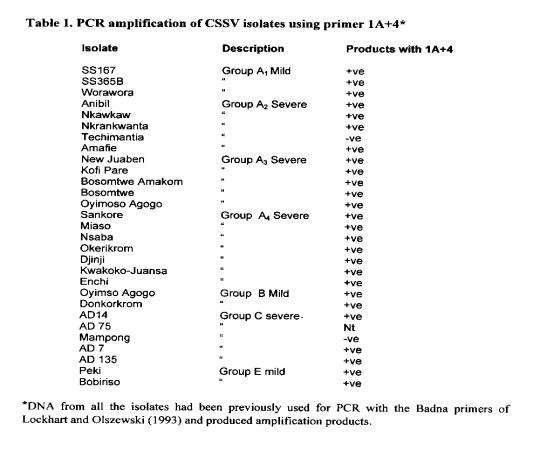
These primers are being screened and in early experiments have produced the expected 600-bp amplification product from 34 of the 36 isolates tested (Table I and Figure 1). The virus DNA samples were extracted from leaves obtained from mature trees, and many of the leaves had no symptoms of infection at the tIme of sampling.
A field survey has been planned to screen these primers further. However, these primers could be made more universal if sequence data from CSSV isolates from other countries could be incorporated. It must be pointed out also that these primers only detect the bacilliform, double stranded DNA genome, and would not detect infections by virus such as the spherical cocoa necrosis virus described by Adomako et at (1974). The main objective of these studies is to provide a sensitive, specific diagnostic system for CSSV indexing.
Inoculation systems
Application of virus inoculum for challenging new progenies is a critical part of the resistance-breeding programme. The objective is to apply the same amount of the pathogen to the seeds/seedlings in order to evaluate the relative impact on them. For CSSV, the current methods include grafting, mealybug transmission and mechanical inoculation using virus purified from infected tissue. Regarding the first two methods, the amount of inoculum applied cannot be determined. While mechanical inoculation is predisposed to being more empirical, the infectivity of highly purified virus tends to be
erratic. Consequently, there remains a need for quantitative methods for inoculum delivery
One of the approaches being currently developed is the use of full-length infectious clones. Infectious virus DNA inoculum can be delivered. using a gene ciun/biolistic system, or by Agrobacterium-mediated transfection of seedlings. Both approaches have been successfully applied in two main initiatives, at CIRAD/INRA in France, and at CRIG Tafo, in Ghana. In Ghana, the preferred method is particle bombardment in a biolistic system, which does not require a biological safety and control facility for monitoring Agrobacterium. Currently several full-length clones of both severe and mild isolates are being evaluated. One mild strain and one severe strain have been shown to be infectious (Figure 2). The latter has been compared to the native virus from which it was cloned, and shown to be identical in its properties: particle composition, symptoms induced, disease severity and transmission by Planococcoides njaIensis~ the preferred vector.
The use of infectious full-length clones for screening for disease resistance has a number of advantages:
The current CRIG programme in this respect is therefore to clone as many isolates as possible from different cocoa growing districts. These would be compared by RFLP and nucleotide sequencing to determine the relationships between them. Their biological effect on the Amelonado host, the susceptible positive control, would also be evaluated before use. Virus clones can, in theory, be selected for screening of cocoa progenies for different areas based on the predominant virus form in those areas,
Tissue culture
Another technology that could benefit breeders in this regard is tissue culture, particularly somatic embryogenesis. Tissue culture potentially can provide breeders with the means to rapidly multiply suitable cocoa clones. It can also be combined with new methods for inoculum delivery (particle bombardment and Agrobacterium mediated transfection) to screen progenies or clones. When beans/seedlings are screened, the number of samples and space required can be limiting. Screening of somatic embryos eliminates these limitations, allowing for large numbers of clones and replicates to be screened. It is furthermore of interest to study the possibility, as with zygotic embryos, that the virus might be eliminated in the somatic embryogenesis process. This would be very useful to facilitate intermediate quarantine procedures.
Conclusion
In conclusion, there are many new emerging technologies that may be applied to various breeding programmes for cocoa improvement. The key to current research is increased collaboration between institutions and scientists. Such collaboration, among other benefits, results in a wider range of pathotypes for the development of assay systems and sharing of information related to new techniques. Thus, for example, access to CSSV isolates from other countries in the West African sub-region would enable development of universal diagnostic system for detection of all Badnaviruses of cocoa in the region where that pathogen is endemic.
Acknowledgements
Funding support for molecular cloning, PCR and biolistic inoculation was from American Cocoa Research Institute, and additional support for PCR research was from Ghana Cocoa Growing Research Association of the UK.
References
Adomako D., DE Lessemann and H.L. Paul. 1983. Improved method for the purification and detection of cocoa swollen shoot virus Ann AppI BioL 103: 109-i 16

Biotech Glossary |
Bioinformatics |
Lab Protocol |
Notes |
Malaysia University |
Novel Technologies for Disease Indexing and Screening for CSSVD Resistance
Sammy T. Sackey
Department of Biochemistry, University of Ghana, Legon, Accra Previous affiliation: Cocoa Research Institute of Ghana (CR10), New Tafo Akim, Ghana
Adomako D. G. K. Owusu and K. K. Oduro. 1974. Purification of cocoa necrosis virus from cocoa leaves. Phytopathology 64: 1325-1330.
Anonymous 1936a Report of the Agronomy Section, West African Cocoa Research Institute, Tafo Station. Ghana.
Anonymous. 1 936b A new disease of cocoa in the Gold Coast. Gold Coast Farmer 5: 122
Brunt A.A, RH. Kenten and H.L. Nixon. 1964. Some properties of cocoa swollen shoot vfrus. J.Gen. Microbial. 36: 303-309.
Hagen L.S., M. Jacquemond, A. Lepingle, H. Lot and M. Tepfer. 1993. Nucleotide sequence and genomic organisation of cocoa swollen shoot virus. Virology 196: 619-628.
Hoffmann K., ST. Sackey, H.J. Vetten, E. Maisse and 0. Adomako. 1997. Immunocapture polymerase chain reaction for the detection and identiflcation of cocoa swollen shoot virus 1A isolates. J. Phytopathology 145: 205-212.
Hughes J. d’A. and L.A.A. Ollennu. 1993. The virobacterial agglutination test as a rapid means of detecting cocoa swollen shoot virus. Ann. AppI. Biol. 122: 299-310.
Lockhart B.E.L. 1990. Evidence for a double stranded DNA genome in a second group of plant viwses. Phytopathology 60: 127-131.
Lockhart BE. and N. Olszewski. 1993. Serological and genomic heterogeneity of banana streak Badnavirus: Implications for virus detection in Musa piants. Pages 105-113 in Breeding Banana and Plantain for resistance to diseases and pests. (Ganry, J.. ed) Proceedings of the International Symposium on Genetic improvement of bananas for resistance to diseases and pests. 7-9 September 1992. Montpellier, France. CIRADIINIBAP.
Lot f-I., E. Djiekpor and M. Jacquemond. 1991. Characterisation of the genome of cacao swollen shoot virus. J. Gen, Virol. 72:1735-1739.
Ollennu L.A.A.., G.K. Owusu and J.M. Thresh. 1989. The cocoa swollen shoot virus eradication campaign in Ghana, Cocoa Growers’ Bulletin 42: 25-35.
Sackey ST., F. Osae-Awuku and M. Manu-Boafo. 1990. Pages 145 in Annual Report of the Cocoa Research Institute of Ghana, CRIG, Accra, Ghana.
Sackey 5.1, ST. Lowor, H.D-Obiatey, G.K. Owusu, 0. Adomako, K. I-loffmann, H.J. Vetten and E. Maiss. 1995, Polymerase chain reaction and nucleic acid hybridisation methods for the detection and differentiation of cocoa swollen shoot virus isolates, Pages 191-200 in Proceedings of First International Cocoa Pests and Diseases Seminar, 6-10 November 1995, Acera, Ghana.
Sackey ST., N.E. Olzewski and B.E.L. Lockhart. 1998. (In press) Isolation of CSSV genomic species by molecular cloning: prospects for application to mild strain cross protection, In Proceedings of the 2B~ International Cocoa Pests and Diseases Seminar, 19-24 January 1998! Yamoussoukro, Cote d’Ivoire.
Sagemann W., H.L. Paul, D. Adomako and O.K. Owusu. 1983. The use of enzyme linked immunosorbent assay (ELISA) for detection of cacao swollen shoot virus (CSSV) in Theobrorna cacao. Phytopath. Z. 106: 281-284.
Sagemann W., 0.E. Lesemann, H.L. Paul, 0. Adomako and G.K. Owusu. 1985. Detection and comparison of some Ghanaian isolates of cacao swollen shoot virus (CSSV) by enzyme linked immunosorbent assay (ELISA) and immunoelectron microscopy (IEM) using an antiserum to CSSV strain 1A.. Phytopath. Z. 144: 79-69.
Thresh J.M. 1980. The origins and epidemiology of some important plant virus diseases. Ann. AppI. Biot, 5: 1-65.
Thresh J.M. 1991. The ecology of tropical plant viruses. Plant Pathology 40: 324-339,
Thresh J.M. and G.M. Owusu, 1986. The control of Cocoa swollen shoot disease in Ghana: an evaluation of eradication procedures. Crop Protection 5: 41-52.
Thresh J.M., G.K. Owusu and L.A. Ollennu, 1988. Cocoa swollen shoot: an archetypal crowd disease, J. Plant Disease 95: 428-446