Molecular genetic markers provide, among other applications, the opportunity to verify identity in germplasm collections. DNA-based polymorphisms are a powerful tool in genetic characterisation. RFLP markers were the first to be used for plant genome studies, for mapping and for diversity analyses. However, RFLPs are labour intensive, time consuming, require a large quantity of DNA and purification by ultra centrifugation. The FCR based techniques, including microsatellite analysis, require less DNA than RELP markers and are therefore convenient for genetic analysis on young plants. An additional advantage of the use of microsatellites is the codominant mode of inheritance, which in contrast to the dominant PCR markers based on arbitary primers, allows easy transfer of markers between genetic maps of different crosses. Compared to RFLPs, microsatellites detect more alleles and a higher level of polymorphism and are equally powerful tools for estimation of heterozygosity. Results obtained on cocoa accessions included in the CFC/ICCO/IPGRI project on Cocoa Germplasm Utilization and Conservation, a Global Approach’ are presented here, including about 150 comparisons of DNA samples from nine collections concerning 28 different accessions. It is concluded that identification problems occur frequently, on average in about 30% of the samples examined in our study. These identification problems occur both in comparisons between accessions from different collections and between trees within accessions from the same collection. Such identification problems constitute a serious problem for comparative analysis of clones obtained from different sites or even from different trees within the same accession. Convenient loci number for identification analyses using microsatellites, sample collection methodology as well as conditions required for comparisons between different microsatellite analyses or between laboratories are briefly discussed.
Introduction
An important consideration when conducting experiments in several different countries is the need to ensure that the experimental material being used in all the sites is correctly identified. Mislabelled material can seriously compromise the interpretation of the data generated. The objective of this study was to verify the conformity of clones used in the CFC/ICCO/IPGRI project on Cocoa Germplasm Utilization and Conservation: a Global approach’ (Eskes et at 1998). Among other activities, this project supports the establishment of an International Clone Trial in ten cocoa producing countries.
PCR-based techniques are widely used to detect polymorphism in plants. Microsatellite markers can detect a great number of alleles that enable discrimination between even closely related individuals. Microsatellite polymorphisms have been successfully used for population and pedigree analyses (Plaschke et at 1995, Provan et at 1996, Senior et at,1998), for genetic mapping (Ramsay et at 2000, Ternnykh et aL 2000) and genotype identification (Rongwen et al. 1995). They were found to be more polymorphic than RFLF markers in rice (Wu and Tanksley 1993) or Arabidopsis (Bell and Ecker 1994). They also appeared to be a good tool for detection of adulteration and mixing in seed samples or in food (Bligh 2000).
In our study, eight microsatellite markers were used to study the identity of 28 ‘International Clones’ used in the CFC/ICCO/IPGRI project that were multiplied in the
quarantine faciJities of The University of Reading, UK and of CIRAD in Montpellier, France. DNA samples were obtained to compare these genotypes with similarly named clones represented in the germplasm collection of origin (source collection’) and in collections in other countries where the International Clone Trial will be established (Eskes et a!. 1998). The microsatellites selected for this study were mapped on eight different cocoa chromosomes (Risterucci ot aI. 2000) (see also Table 1), A comparison of the heterozygosity level obtained by RFLP and microsatellite studies is reported here as well for twenty cocoa genotypes.
Materials and methods
Plant material
DNA isolation
Microsatellite analysis
Results and discussion
Use of microsatellites for identity studies
It is to be noted that when the DNA profile is different for at least one locus one can conclude that the genotypes tested are different. However, when the profiles are similar, there is no absolute guarantee that the genotypes are really identical, It is known that genetically related genotypes can show identical DNA profiles but have some morphological differences (such as fruit colour or shape).
It is concluded that accessions that have been transferred between cocoa gerrnplasm collections may frequently be subject to errors in identification. Differences are present between collections as well as between trees within one collection. These differences were established with regard to reference samples, in this case obtained from one plant of the accessions maintained in intermediate quarantine facilities in Reading and Montpellier. The present study does not permit us to judge, in case of differences, which of the accessions are to be considered as the true original ones. Mistakes in labelling or identification may have occurred in the source collection as well as in recipient collections.
Estimation of the level of heterozygosity
In our study, we compared RFLP and microsatellite markers to estimate the level of heterozygosity of twenty cocoa clones (Table 4). For RFLPs, 29 to 33 loci were used and for microsatellites 19 loci. For these clones, the average level of heterozygosity obtained was 22.9 % for RFLPs and 45.6 % for microsatellites. As expected, the higher degree of allelic diversity and polymorphism of microsatellite loci compared to genomic or cDNA RFLPs shows that microsatellites may constitute a befter tool for comparisons of the degree of heterozygosity.
General discussion
Further development and use of microsatellite markers
Choice of microsatellite loci for identity control
Number of alleles
Recommendations for leaf sample collection and shipment
Codification method to permit comparison between different analyses and laboratories
Construction of a database for microsatellite alleles for cocoa
Acknowledgements
References
Bell C.J. and J.H. Ecker. 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137-144.

Biotech Glossary |
Bioinformatics |
Lab Protocol |
Notes |
Malaysia University |
Use of Microsatellite Markers for Germplasm Identity Analysis in Cocoa
Cirad-Amis
TA 40/03, Avenue Agropolis, 34398 Montpellier Cedex 5, France
Fresh or dried leaves were sent in 1998 and 1999 to Montpellier from Reading and from the nine collections in cocoa producing countries (see Tables 2 and 3). Most of these clones form part of the so-called ‘International Clone Trial’ in the CFC/ICCO/IPGRI project (Eskes of at 1998).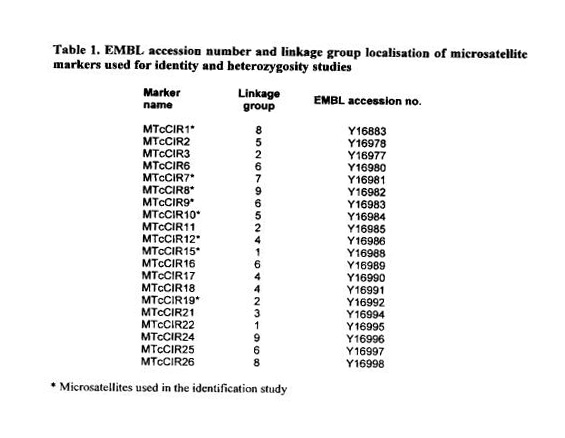
A new DNA isolation protocol was used to obtain the small quantities of DNA needed for microsatellite and AFLP analyses. DNA was usually isolated from fresh adult leaves. One gram of tissue, frozen in liquid nitrogen and powdered with a mortar, was mixed with 5 ml of extraction buffer (1.4 M NaCI, 100 mM Tris HCL pH 8.0, 20 mM EDTA, 10 mM Na2S03, 1 % PEG 6000, 2% MATAB) preheated to 75°C. After homogenisation for 10 s with a vortex, the extract was incubated for 30 mm at 75°C. After cooling to 20°C, an equal volume of chloroform-isoamyl alcohol (24:1 v/v) was added, followed by emulsification. The tube was then centrifuged at 7000 g for 30 mm and the supernatant was precipitated at —20°C overnight after addition of an equal volume of isopropanol. The DNA was removed with a glass hook and re-suspended in 1 ml of TE.
A genomic library enriched in simple sequence repeats was constructed for microsatellite—analysis using a modified version of the protocol of Karagyozov et a!. 1993 (Lanaud et at 1999). The primers were end labelled with y-33P ATP, and amplification was performed in a MJ Research PTC 100 Thermal cycler on 20 pl reaction mixtures containing 10 ng of cocoa DNA, 0.2 mM dNTP mix, 2 mM MQCI2, 50 mM KCI, 10 mM tris-HCI (pH 8.3), 0.2 pM primer (5’ end labelled with y-33P ATP) and 1 unit of Taq polymerase (Eurobio). The samples were denatured at 94°C for 4 mm and subjected to 32 repeats of the following cycle: 94°C for 30 s, 46°C or 51°C for 1 mm and 72°C for 1 mm. After adding 20 pl of loading buffer (98% formamide, 10 mM EDTA, bromophenol blue, xylene cyanol), the mixes were denatured at 92°C for 3 mm and 3 p1 of each sample were loaded onto 5% polyacrylamide gel with 7.5 M urea and electrophoresed in 0.5% TEE buffer at 55 W for 1 h 40 mm. The gel was dried for 30 min at 80°C and exposed overnight to X-ray film (Fuji RX). EMBL accession numbers of the microsatellites used are listed in Table 1.
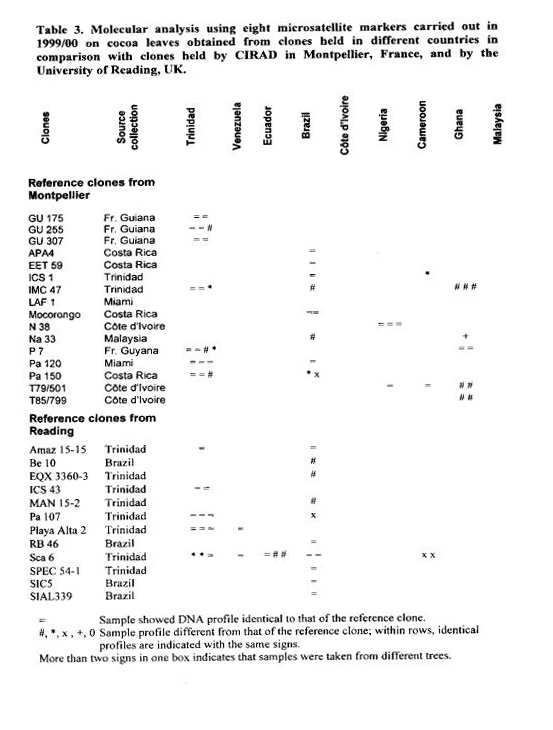
In the first round of analyses, in 1998/99, 75 DNA samples obtained from clones in nine collections in cocoa producing countries were compared with the 28 samples from the reference clones, 13 from the Montpellier and 15 from the Reading greenhouses (Table
2). The results showed differences in profile, for at least one of the eight microsatellite loci, with regard to the reference clones for 19 out of the 75 comparisons (25%). For 13 out of the 28 clones (46%) in at east one of the samples tested differences were found with regard to the reference samples.
New analyses were performed in 1999/00 (Table 3) comparing 12 clones from Reading and 16 from Montpellier with samples received from seven countries. The objective was to confirm data obtained in the first year and also to carry out new comparisons. Seventy-six DNA samples were compared to the 28 reference samples. This time, 27 samples (36%) showed differences in profile for one or more of eight microsatellite loci in relation to the reference clones, Twelve out of the 28 clones (43%) showed differences for at least one of the DNA samples compared.
For both years it was observed that misidentifications could also be frequently found amongst the trees within an accession. This was the case for eight out of the 23 accessions (35%) for which more than one tree was tested from the same accession.
Comparisons of identity between years are limited to the samples of the reference clones and of clones with similar profile. Samples that were different from the reference clones in the two years have not been compared together in one analysis to verify if the differences found could relate to the same genotype.
An interesting result concerns the widely distributed SCA 6 clone. The samples analysed from the nine collections indicate that the identity of this clone is wrong for all four samples from Cameroon, for one of the two introductions in the collections in Ecuador (Loma Long’) and for two trees out of four trees tested from the Trinidad collection. Another intriguing result is that the sample from the reference clone can be different fro m the sample received from the source collection’, i.e. in the country of origin of the clone (e.g. BE 10 and MAN 15-2 from Brazil).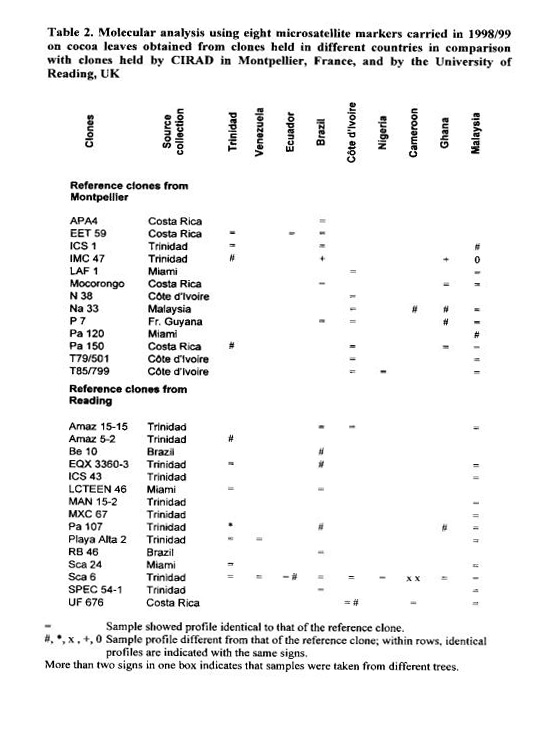
Molecular markers have been used to estimate the genetic diversity and other parameters which can be useful in the choice of genotypes for breeding purposes (NGoran et at 2000). Co-dominant markers permit breeders to compare the level of heterozygosity between candidate genotypes for breeding. Homozygous parents are desirable for the production of uniform hybrids, whereas more heterozygous parents may be more useful in creating new recombinations for clone selection.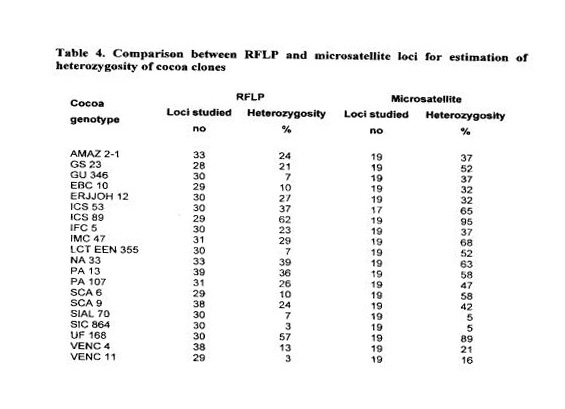
With microsatellites, rapid screening of large numbers of plants may become feasible. Although their development is costly and time consuming, microsatellites are a good choice for many types of genome study, such as an internationally co-ordinated effort to genotype cocoa germplasm. Presently. 69 mapped microsatellites are available for cocoa. The production of more microsatellites with the French National Sequencing Center (CNS Evry) and subsequent screening of these microsatellites at CIRAD will continue.
The choice of loci on different chromosomes appears to be an important criterion in obtaining good coverage of the genome. Other important criteria are the allelic frequencies and the level of polymorphism that can be identified for each locus. By choosing the most suitable microsatellite loci in this way, 8 to 9 loci each with 6 to 10 alleles would appear sufficient for an initial verification of identity; nevertheless the use of 15 loci would probably be necessary for safe identification of more closely related genotypes.
In the future, to allow more widespread and easier use of microsatellites, markers will be selected that can give results without the use of radioactivity, i.e. by using agarose gel with BET staining or a small acrylamide gel with silver staining.
The efficiency of genotype identification with different kinds of molecular markers depends on the number of alleles that can be identified at a single locus. Comparisons between RFLP and microsatellite markers on cocoa genotypes showed on average 2.4 alleles per locus for RFLPs and 5.6 alleles per locus for microsatellites. Similar results were found in others species, 2.2 and 4.3 for soybean (Morgante et a!. 1994), 2.6 and 5.4 for barley (Russel et a!. 1997), and 2.5 and 7.4 for rice (Olufowote et a!. 1997), respectively.
One leaf in good condition is a large enough sample for DNA extraction and microsatellite analysis. Fresh leaves conditioned in slightly humidified paper that were sent to the Montpellier laboratory for the identity studies gave good DNA extraction results if the transit time did not exceed two weeks. If it is not possible to use fresh leaves, air-dried leaves (not oven-dried leaves) or extracted DNA samples can be used as good alternatives.
It is necessary for all laboratories to have a similar method for allelic codification of microsatellite loci. The best method would seem to be to codify in relation to reference alleles of a homozygous clone, e.g. Catongo. DNA of such a clone should be made
available to all laboratories in order to allow for comparisons relative to the reference allele.
A further requirement for more frequent comparative use of microsatellites in cocoa genome analyses is the construction of an international database for microsatellite loci, containing all information availabie and permitting easy comparisons with analyses carried out elsewhere.
The research on identity of cocoa clones presented here is partially supported by the
CFC/ICCO/IPGRI Project. This project is developed under the aegis of the International
Cocoa Organization and mainly funded by the Common Fund for Commodities. The
supply of leaf samples for DNA extraction by the following institutions is acknowledged:
CEPLAC in Brazil, CNRA in Cote divoire, CRIG in Ghana, CRIN in Nigeria, CRU in
Trinidad, INIA in Venezuela, JNIAP in Ecuador, IRAD in Cameroon, MCB in Malaysia and
The University of Reading in the UK.
Bligh H.F.J. 2000. Detection of adulteration of Basmati rice with non-premium long-grain rice. International Journal of Food Science and Technology 35: 257-265.
Eskes AB., J.M.M. Engels. and R.A, Lass. 1998. The GFC/ICCO/IPGRI Project: a new initiative on cocoa germplasm utilization and conservation. Plantations Recherche Developpement 5: 412-422.
Karagyozov L., I.D. Kalcheva and V.M. Chapman. 1993. Construction of random small-insert genomic libraries highly enriched for simple sequence repeats. Nucleic Acids Research
21: 3911-3912.
Lanaud C., AM. Risterucci, I. Pieretti, M. Faique, k Bouet and P.J.L. Lagoda. 1999. Isolation and characterization of microsatellites in Threobroma cacao L. Mol Ecol 8:2141-2152.
Morgante M., A. Rafalsici, P. Biddle, S. Tingey and AM. Olivieri. 1994. Genetic mapping and variability of seven soybean simple sequence repeat oci. Genome 37: 763-769.
N’Goran J.A.K., V. Laurent, AM. Risterucci and C. Lanaud. 2000. The genetic structure of cocoa populations (The obroma cacao L,) revealed by RFLP analysis Euphytica 115: 83-90.
Olufowote JO., V. Xu, X. Chen, W.D. Park, H.M. Beachell, RH. Dilday, M. Goto and S. R. Mccouch. 1997. Comparative evaluation of within-cultivar variation of rice (Oryza sativa L.) using microsatellite and RFLP markers. Genome 40: 370-378
Plaschke 3., MW. Canal and MS. ROder 1995. Detection of genetic diversity in closly related bread wheat using microsatellite markers. Theor, AppI. Genet. 91:1001-1007.
Provan J., W. Powell and It Waugh. 1996. Microsatellite analysis of relationships within cultivated potato ( Solanum, tuberosum) . Theor. AppI. Genet. 92: 1078-1 084.
Ramsay L., M. Macaulay, S. Degli Ivanissevich, K. MacLean, L. Cardle, J. Fuller, K.J. Edwards,
S. Tuvesson, M. Morgante, A. Massari, E. Maestri, N. MarmiroJi, 1. Sjakste, It Ganal, W.
Powell and R. Waugh. 2000. A Simple Sequence Repeat-Based Linkage Map of Barley.
Genetics 156: 1997-2005.
Risterucci AM., L. Grivet, J.A,K. N’Goran, I. Pieretti, M.H. Flament and C. Lanaud. 2000. A high-density linkage map of Theobroma cacao 1. Theor. AppI. Genet, 101: 948-955.
Rongwen .J., MS. Akkaya, A.A.. Bhagwat, U, Lavi and P.B.Cregan. 1995. The use of microsatellite DNA markers for soybean genotype identification. Theor. AppI. Genet. 90: 43-
48.
Russell JR., JO. Fuller, M. Macaulay, B.G. Hatz, A. Jahoor, W. Powell and R. Waugh. 1997. Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs, Theor. AppI. Genet. 95: 714-722.
Senior ML., J.P. Murphy, MM. Goodman and OW. Stuber. 1995. Utility of SSRs for
determining genetic similarities and relationships in maize using an agarose gel system. Crop Sci 38: 1088-1098.
Temnykh S., W.D. Park, N. Ayres, S. Cartinhour, N. Hauck, L. Lipovich, Y.G. Cho, 1. lshii and SR. McCouch SR. 2000. Mapping and genome organization of microsateilite sequences in rice (Oryza sativa L.). Theor. AppL Genet. 100: 697-712.
Wu KS. and S.D. Tanksley. 1993. Abundance polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet. 241: 225-235.