Living germplasm collections of Theobroma cacao genotypes are maintained in several international collections scattered throughout Central and South America as well as in selected Caribbean Islands such as Puerto Rico and Trinidad and Tobago. These living germplasm Collections predate the current curators and the scope of the genetic diversity within these collections has never been determined in detail. Preliminary molecular studies on a large international collection in Trinidad and Tobago indicate that as much as 20-30% of the collection may be mislabelled or labelled with different names. The United States Department of Agriculture has begun a programme to identify and describe the genetic diversity of these collections using state-of-the-art molecular fingerprinting techniques. Two separate molecular analysis techniques, Amplified Fragment Length Polymorphism (AFLP) DNA analyses and Simple Sequence Repeats (SSR or microsatellite) DNA analysis were performed on populations of trees in T. cacao germplasm collections to evaluate the utility of these procedures for DNA fingerprinting of this tree crop. DNA fragments were selectively amplified, labelled with fluorescent dyes and separated by capillary electrophoresis using two different models of DNA analysers (an ABI/Perkin Elmer 310 single capillary injector and a Beckman CEQ 2000 eight channel capillary DNA analyser). Using either procedure, electropherogrammes of DNA fragment patterns were reproducible and consistent within a common genotype, while differentiating separate genotypes. Similarity dendrograms were based on the combined cluster analysis of AFLP primer sets of polymorphic peaks or from SSR primers selectively amplified with PCR technology. Based on this study, 15 primers for SSR markers have been selected as an international standard technique for 7 cacao molecular characterisation. Molecular fingerprinting defines individual accessions identifies duplications of genotypes within germplasm collections, corrects mislabelling of accessions and estalishes genetic similarity of breeding lines for potential crosses. This will help also to select for germplasm stocks to be used in breeding Theobroma cacao cultivars with improved tolerance/resistance to diseases.
Introduction
In the last 15 years, the rapid spread of three major fungal diseases affecting Theobroma cacao together with unstable prices for cocoa beans, has reduced production of the commodity in Central and South America and nearby Caribbean Islands by more than 75% of former values. The three most important diseases that threaten cocoa production in the Americas are Witches’ Broom disease caused by Crinipellis perniciosa, Frosty Pod Rot caused by Monillophthora roreri and Black Pod Rot caused by several species of Phytophthora. The development of genetically resistant 71 cacao is one technique that is being employed to help combat this problem. However, the germplasm collections available as genetic resources are comprised of cocoa accessions which have never been fully characterised, and the information about the genetic diversity within each collection is lacking.
Furthermore, it seems likely that many identical accessions in the collections have been given different names due to the dates and conditions of collection. In a preliminary study in Trinidad and Tobago, using RAPD molecular markers, it was estimated that up to 20% of the plots could contain misidentified material due to errors during germplasm transfer or long- term maintenance (Christopher et al. 1999).
To assist in the development of genetically resistant cultivars to these diseases in Theobroma cacao, the United States Department of Agriculture (USDA) has begun a multi-faceted genomics programme focused on T. cacao. The programme is comprised of integrated efforts at three USDA locations that include:
This report will focus on the establishment of the International Cocoa Molecular Identification Centre in Beltsville, and the activities of this programme on the characterisation of 71 cacao germplasm collections by DNA fingerprinting procedures.
Material and methods
DNA isolation
AFLP DNA analysis
SSR DNA analysis
Results and discussion
Selection of DNA fingerprinting techniques
AFLP DNA analysis
The use of AFLP DNA analysis on T. cacao involves five basic steps for molecular analysis. First the genomic DNA is isolated from the plant leaf and thoroughly fragmented using the frequent cutting restriction enzymes EcoRl and Msel. These restriction enzymes completely digest the genomic DNA into fragments of approximately 50 — 1000 bp during a complete overnight digestion. The fragmented DNA is then ligated with a small segment of known DNA and selected fragments of the DNA are copied by PCR amplification. In the third step of the process, further specificity is achieved by a second, selective amplification in which the PCR primers are modified to amplify only a fraction of the genome. In the fourth step of the process, these fragments must be separated to produce the actual fingerprint upon which subsequent analysis is accomplished. At present, either electrophoresis on slab gels or capillary systems are used for the separation of the DNA fragments. The final step in the AFLP process is the scoring and data analysis of the DNA fragments. The scoring of the DNA fragments is linked to the separation system and involves the need to visualise the DNA fragments by staining, fluorescent dyes, or with radioactive isotope markers. Each of these DNA visualisation procedures brings its own set of criteria for accurate scoring and analysis of the data that is unique to that DNA marker.
One of the main advantages of the AFLP DNA analysis technique is that it is not necessary for the researcher to have previous knowledge about the genome being tested in order to achieve results capable of differentiating between genotypes. Having said that, however, all AFLP primers do not produce equally valuable information about
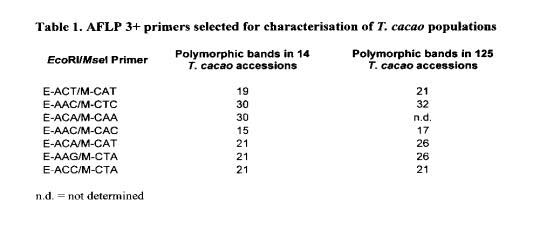
the nature of the DNA being tested. Many AFLP primers work better in some plants than in others. To optimise AFLP techniques for T. cacao, we evaluated the performance of 36 different combinations of AFLP primers from EcoRl and Msel double restriction enzyme digests having three selected base pairs on each primer. From these primer combinations the seven primers shown in Table 1 were used to generate singular and composite comparative dendrograms for two differently sized populations of T. cacao containing 14 and 125 accessions respectively. These two population sizes were chosen to evaluate the usefulness of the AFLP DNA analysis technique on different sample sizes. There are several parameters that would define optimal primers for use by the AFLP analysis techniques. Optimal primers produce easily scorable DNA fragments that show reproducibility between samples and a high number of polymorphic bands to elucidate diversity and discriminate between samples.
AFLP DNA analysis produces 70 to 90 DNA fragments per primer for each accession of T. cacao tested. Although many of these bands are conserved in nature, that is they are present in all of the samples that are analysed, a large number of the bands are polymorphic in comparison to the rest of the sample in the population being tested. In our hands, we declared a band polymorphic if it was absent in at least one of the other accessions being evaluated in that population and could be reliably separated and scored as a single band. Thus the number of polymorphic bands recorded in Table 1 is a conservative estimate of the total number of polymorphic bands produced. However, these could be repeatedly scored with accuracy. On average, there were usually 20-30 polymorphic bands per accession using the primers that we selected for optimal performance.
As Table 1 shows, the number of polymorphic bands is also related to the genetic diversity of the population being examined and the number of samples in the population. In all but one primer, the number of polymorphic bands increased with the sample size, and thus the diversity of the samples increased. The number of scorable bands for AFLP is quite high in comparison to RAPD and RFLP procedures where 1-4 bands per probe would be considered substantial (see Lin eta!. 1996).
The question arises as to how many primers are sufficient to adequately predict the genetic diversity of a population. If the population is very small, and the goal of the analysis is to determine if the samples are closely related, then one or two primers may be sufficient to predict the relationships among the samples. Larger populations of samples may require more in-depth analysis with higher numbers of primer pairs. To determine the optimal number for any population size, a dendrogram was produced after the analysis of each primer pair and composite dendrograms were complied ri a stepwise fashion for each primer to determine the degree of reliability of the data with additional primer analyses. When additional primers did not significantly change the pattern of the dendrogram, then a sufficient number of analyses had been done for that sample size.
SSR DNA analysis
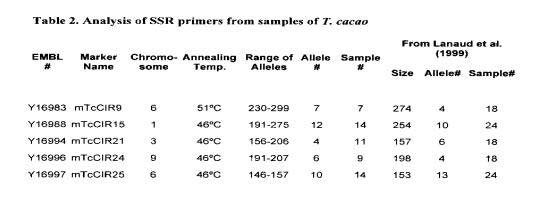
Table 2 shows the data obtained from an examination of five SSR primers selected from four different chromosomes across the cocoa genome. A population of up to 14 I cacao accessions was subjected to PCR amplification based on these primers and the numbers of individual allelic markers were recorded for each of the primers. These data closely parallel previously published reports of Lanaud eta!. (1999) using different T. cacao accessions. This close correlation demonstrates the utility of various laboratories using SSR primers as reproducible markers for DNA fingerprinting studies of this type. The results also show that these SSR loci have a high degree of variability associated with this population of plant samples, allowing for assignment of a unique molecular description of each of the samples with three to four SSR primers.
Conclusions
We have examined two different molecular analytical systems, namely AFLP and SSR DNA analysis, for their utility as a procedure for the molecular characterisation of international germplasm collections of I cacao. Both techniques are useful and can be used to clearly define the genetic identity and diversity of these valuable resources. Due to the ease of data analysis, reproducibility, and simplicity of the procedural operations, the use of SSR DNA analysis has been recommended as an international standard for T. cacao. Further, 15 specific SSR primers will be selected as the international standard primers for SSR analysis of T. cacao so that international agreement of nomenclature can be applied uniformly throughout the global cocoa community.
References
Christopher Y., V. Mooleedhar, F. Bekele and F. Hosein. 1999. Verification of accessions in the ICG,T using botanical descriptors and RAPD analysis. Pages 15-18 in Annual Report of the Cocoa Research Unit for 1998. Cocoa Research Unit, The University of the West Indies. St. Augustine, Trinidad and Tobago.

Biotech Glossary |
Bioinformatics |
Lab Protocol |
Notes |
Malaysia University |
USDA DNA Fingerprinting Programme for Identification of Theobroma Cacao Accessions
James A. Saunders’, Alaa A. Hemeida2 and Sue Mischke’
1 - USDA, Agricultural Research Service, Bldg. 50, Rm. 100, Beltsville, MD 20705, USA
2 - Genetic Engineering & Biotechnology Research Institute (GEBRI), Sadat City, Minufiya University, Egypt
DNA isolations were performed on 100mg samples of frozen leaf tissue from 71 cacao accessions collected from either the USDA Tropical Agricultural Research Station in Mayaguez, Puerto Rico or the CEPLAC Research Institute, Itabuna, Bahia, Brazil. Tissue disruption was accomplished with one 40-second shaking regime on a BiolOl FastPrep (Carlsbad, CA) rapid oscillating shaker fitted with 2 ml tubes containing garnet and a single 550 mg ceramic bead, For lack of a better name this “shaker/basher” produces a fine homogenate of leaf material by the abrasive actions of the garnet and ceramic bead striking against the leaf material while the assembly is being rapidly shaken in the presence of extraction buffer. This procedure has the distinct advantage over grinding with a mortar and pestle in that 12 samples can be processed simultaneously with no possibility of cross contamination of leaf material since the samples are completely contained within labelled, disposable tubes, Purified DNA samples were isolated from this material using a modified Dneasy Plant Mini DNA isolation kit by Qiagen (Hilden, Germany). Double stranded DNA was quantified by the PicoGreen fluorescence technique (Molecular Probes, Inc., Eugene, OR) using a Fluoroskan Ascent (Labsystems, Helsinki, Finland) microplate reader equipped with 485/538 excitation/emission filter settings.
Techniques for the AFLP analysis of I cacao were adapted and modified from those published previously (Lin et aL 1996; Saunders et al. 2000, in press). The modifications consisted of the use of fluorescent dyes linked to primers in pJace of 32P, and the use of capillary electrophoresis for DNA size fragment separation.
Two different capillary electrophoresis systems were used to evaluate the DNA fragment analysis. These systems consist of either an ABI Prism Perkin Elmer 310 single-channel capillary Genetic Analyser (PE Applied Biosystems, Foster City, CA) used for AFLP DNA analysis or a Beckman CEQ2000 eight-channel capillary DNA Analysis System (Beckman Coulter, Fullerton, CA) used for SSR DNA analysis.
For the ABI PE 310 capillary DNA fragment analyser, 5.5p1 containing 50-200 ng of genomic DNA from each sample was digested overnight at room temperature and ligated to adaptor pairs of known sequence following manufacturer’s instructions from the AFLP Plant Mapping Kit (PE Applied Biosystems, Foster City, CA). The ligation pairs and digestive enzymes were contained within an additional 5,5~&l of reagent mixture of 1 unit/jil EcoRl and 1 unit/pi of Msel restriction enzymes, 50 mM Tris-HCI pH 7.5, 10mM MgCI2, 10mM dithiothreitol, 1 mM ATP, 25 1sg/ml BSA, 60 Units of T4 DNA ligase (New England Biolabs, Beverly, MA), 1 ii of each EcoRl and Msel adaptor pairs (AFLP Plant Mapping Kit, PE Applied Biosystems, Foster City, CA), 0.05 M NaCI and 0,6 ~xl sterile distilled water. After the restriction-ligation of the DNA, the samples were diluted 18-fold with 15 mM tris-HCL pH 8.0 containing 0.1 mM EDTA. PCR preselective amplification was performed on the samples with a GeneAmp 9700 PCR system (PE Applied Biosystems, Foster City, CA), in a 20 ~il reaction containing 4 ~il of the restriction/ligated DNA and 16 ~il of a mixture containing 1 jil of EcoRl and Msel AFLP preselective primers (PE Applied Biosystems) and 15 jil of AFLP core mix (PE Applied Biosystems). The PCR amplification protocol consisted of 72°C for 3 minutes followed by 20 cycles of the following profile: 94°C for 20s, 56°C for 30s and 72°C for 2 mm with a final 60°C hold for 30 mm. The amplified product was diluted 20-fold using 15mM tris-HCI buffer pH 8.0 containing 0.1 mM EDTA.
For selective amplification of restriction fragments, seven sets of AFLP selective EooRl and Msel primer pairs (PE Biosystems) were used. The reaction mixture for each selective amplification contained 15 ~xl AFLP core mix, 1 pxl of AFLP EcoRl dye-labelled primer with three additional user selected nucleotides and 1 uI AFLP Msel unlabelled primer with three additional user-selected nucleotides. The PCR profile for the selected amplification consisted of an initial warm-up at 94°C for two minutes then one cycle of 94°C for 20s, 66°C for 30s and 72°C for 2 mm, followed by ten subsequent cycles each with 1°C lowering of the annealing temperature and finally 25 cycles of 94°C for 20s, 56°C for 30s and 72°C for 2 mm with a final hold of 60°C for 30 mm. Following PCR amplification, 1 pi of reaction products, 24 ~tl of deionised formamide, and 0.5 xl Genescan-400 size standard were mixed, heated at 95°C for 5 mm, and stored at 4°C. DNA fragment sizes between 50 and 400 bp were separated and determined on an ABI Perkin Elmer 310 single-channel capillary genetic analyser.
Primer sequences for SSR analysis were described by Lanaud et aI.(1999). For PCR amplification of the SSR primers in Theobroma cacao genomic DNA, five primer sets were synthesised at Research Genetics, Inc. (Huntsville, Alabama). The S’forward primers end-labelled with WelIRed ~ fluorescent dyes were supplied and used according to manufacturer’s instructions (Beckman Coulter Inc., Fullerton, CA, USA). The reaction mixtures contained 3 ~tl genomic DNA (tlOO ng), 1 pi of both forward and reverse primers (final concentration of 0.5 pi , and 15 III of PCR Super Mix (Life Technologies Inc., Gaithersburg, MD). The PCR reaction was performed in a Gene Amp 9700 PCR system with the temperature profile of 94°C for 4 mm followed by 40 cycles of following profile: 94°C for SOs, 46°C or 51°C for mm and 72°C for 2 mm with a final 60°C hold for 30 mm. The individual samples were run on a Beckman CEQ2000 eight channel capillary DNA Analysis system to determine the size of the amplified DNA fragments from each accession of Theobroma cacao. The capillary injection consisted of a 30 second electrophoresis at 2.0 kV from a 40 il mixture of 0.5 psI CEQ2000 DNA size standard-400 (Beckman Coulter P/N 608098), 0.5 p.l of PCR amplification mixture and 39 jil sample loading solution containing deionised formamide (SLS Beckman Coulter PIN 608083). The CEO 2000 Frag-3 profile was used for the running conditions that were; capillary temperature 50°C, denaturation temperature 90°C for 120s, and separation voltage 6.0 kV for a run time of 35mm. Data analysis was performed using the CEQ 2000 Fragment Analysis software according to manufacturer’s recommendations (Beckman Coulter Inc., Fullerton, CA, USA).
Multiple techniques are available for the molecular characterisation of plant germplasm collections with refinements, updates and modifications of these procedures being introduced at an astounding pace. Several researchers have evaluated the use of Random Amplified Polymorphic DNA (RAPD) and Restriction Fragment Length Polymorphism (RFLP) DNA analyses for diversity studies on T. cacao with mixed results (e.g. Wilde eta!. 1992; Gilmour 1994; Laurent eta!. 1994; N’Goran eta!. 1994 and 0. Butler Cocoa Research Unit, Trinidad And Tobago, personal communication). Although both of these molecular analysis techniques produce molecular markers, neither is suitable for the high throughput analysis necessary for the molecular characterisation of the estimated 8,000— 10,000 T. cacao accessions held in present collections within the Americas. In addition, there are questions of consistency and repeatability under the conditions necessary for large-scale projects using RAPD. To address some of these issues, we evaluated two of the more recent DNA analytical procedures, Amplified Fragment Length Polymorphism’s (AELP) and Simple Sequence Repeats (SSR also termed microsatellite analysis) for their utility as DNA fingerprinting procedures for T. cacao germplasm collections. Since the size of the population which is being evaluated was also a factor in the utility of the molecular procedure that is used, we looked at two different T. cacao populations, a small sample of 14 T. cacao accessions from Brazil and a larger population of approximately 125 T. cacao accessions collected from Central and South America, These samples were subjected to DNA analysis using both AFLP and SSR protocols to evaluate the utility of the techniques.
SSR or microsatellite DNA analysis makes use of specific sequences within the genome of T. cacao with a di-, tn-, or tetra-nucleotide repeating unit (Rogstad 1993; Cregan eta!. 1994; Wang eta!. 1994; Cregan and Quigley 1997). In almost all cases, these regions of the genome are non-coding in nature and as such are regions of high genetic variability that are inherited. These areas of repeating nucleotides of the genomic DNA are identified through a variety of sequencing procedures. They can be used for DNA fingerprinting by designing PCR primers based on nucleotide sequence from flanking regions of the genomic DNA. Due to the ease of separation of relatively small fragments of DNA, SSR fragments that range in size from 100—350 bp in length are the most useful for DNA fingerprinting, SSR DNA analysis has several advantages over AFLP DNA analysis since each sample is expected to display a maximum of two peaks or alleles for each unique SSR primer used. These alleles are the result of maternal and paternal inheritance patterns and the size of the alleles and their frequency within a population of plant accessions demonstrate diversity in germplasm being examined. Variability in the size of the fragment occurs due to deletions or insertions in the SSR region of the genomic DNA. It is possible to sometimes get more than two SSR peaks per sample if the DNA is copied in another region of the genome, however with the proper selection of PCR primers these situations can be avoided. Another distinct advantage of SSR DNA fingerprinting is that the quantity and quality of the DNA for analysis is a great deal more forgiving for SSR analysis compared to AFLP procedures. This is primarily due to the fact that the restriction digestion step of the AFLP process is eliminated in the SSR procedure. One distinct disadvantage to SSR fingerprinting is that considerable research has to be accomplished to identify and sequence SSR primers compared to AFLP procedures.
Cregan PB., MS. Akkaya, Ak Bhagwat, U. Lavi and J. Rongwen. 1994. Length polymorphisms of simple sequence repeat (SSR) DNA as molecular markers in plants. Pages 47-56 in Plant Genome Analysis (Gresshoff, P.M., ed). CRC Press, Boca Raton, Florida.
Cregan P.8 and CV. Quigley. 1997. Simple sequence repeat DNA marker analysis. Pages 173-185 in DNA Markers: Protocols, Applications and Overviews. (Caetano-Anolles, G. and Gresshoff, P.M., eds). John Wiley & Sons, New York.
Gilmour M. 1994. The BCCCA ringtest on the RAPD analysis of cocoa. Pages 135-138 in Proceedings of the International Workshop on Cocoa Breeding Strategies, Kuala Lumpur, Malaysia.
Lanaud C., AM. Risterucci, I. Pieretti, M. Faique, A. Bouet and P.J.L. Lagoda. 1999. Isolation and characterization of microsatellites in Theobroma cacao L. Molecular Ecology 8: 2141- 2 143.
Laurent V., A.M Risterucci and C. Lanaud. 1994. Genetic diversity in cocoa revealed by G-DNA probes. Theor. AppI. Genet. 88: 193-1 98.
Lin J.J., J. Kuo, £ Ma, J.A. Saunders, H.S. Beard, M Ft MacDonald, W. Kenworthy, G.N. Ude and BE. Matthews 1996. Identification of molecular markers in soybean comparing RFLP. RAPD and AFLP DNA mapping techniques. Plant Molecular Biology Reporter 14: 156-169.
N’Goran J.A.K., V. Laurent, AM. Risterucci and C. Lanaud 1994. Comparative genetic diversity studies of Theobroma cacao L. using RFLP and RAPD markers. Heredity 73: 589- 597.
Risterucci AM., L. Grivet, JA.K. N’Goran, I. Pieretti, M.H. Flament and C. Lanaud. 2000. A high density linkage map of Theobroma cacao L. Theor. and AppI. Genet. In press.
Rogstad S.H. 1993. Surveying plant genomes for variable number of tandem repeat loci. Methods in Enzymology 224: 278-294.
Saunders J.A., M.J. Pedroni, L. Penrose and T. Fist. 2000. AFLP DNA analysis of opium Poppy. Crop Science. In press.
Wang Z., J.L. Weber, G. Zhong and S.D. Tanksley.1994. Survey of plant short tandem DNA repeats. Theor. and AppI. Genet. 88: 1-6.
Wilde JR., R. Waugh and W, Powell. 1992. Genetic fingerprinting of Theobroma clones using randomly amplified polymorphic DNA markers. Theor. and AppI. Genet. 63: 871-877.